When you order a cDNA plasmid (the DNA copy of the mature messenger RNA (mRNA) instructions for making a protein) from a repository (e.g. addgene, DNASU), it will likely come in some sort of generic cloning plasmid. And you will want to “subclone” it into an expression plasmid if you want to use it to make protein from it (i.e. move it out of the one plasmid and into another). blog form: https://bit.ly/vector_plasmids
There can be a lot of options and weird nomenclature and stuff when it comes to plasmids, but the key things they differ in are usually:
- copy number - how many copies of the plasmid will each cell host (high’s good for cloning, lower for expression)
- promoter usage - how will you tell the cells to use an RNA Polymerase to make mRNA copies of the gene (and subsequently protein from those mRNA instructions) (T7 (w or w/o lac), tac, T5)
- inducible expression of the RNA Pol? (e.g. induce T7 expression with IPTG)
- selection markers (typically antibiotic resistance genes for Amp, Kan, Strep, etc.)
- potentially secretion signals
- epitope tags (His, Flag, etc., which can be at the N-terminus (start of the protein) or C-terminus (end of the protein))
- often with protease cleavage sites (TEV, HRV3C, thrombin, etc.) for removal from protein
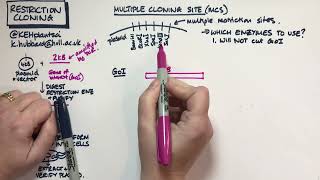
- restriction enzyme cut sites for cloning with restriction enzymes (often there are multiple of them in a region called a multiple cloning site (MCS))
- the different letters after a plasmid number (e.g. pET28a vs pET28b vs. pET28c) usually refer to what reading frame the inserted protein will be read in with respect to the cut site
In terms of reading frames, which plasmid you start with is less of an issue if you’re using a PCR-based cloning strategy (such as SLIC). PCR-based strategies are also good because they let you do “scarless” cloning - you don’t have extra letters on the ends of your proteins that come from having some of the MCS still there. PCR-based strategies are also great because you can easily clone in different tags and things, and do things like swap the tag from the N- to the C-terminus or vise versa, which can sometimes make a difference in terms of tag accessibility, protein folding, etc.. More on cloning methods here: http://bit.ly/molecularcloningguide
plasmids will often have a bunch of notation with things like Δsomething (Δ is delta and it means something is missing, missing things might also be indicated with a superscripted - sign)
- for example, recA- cells are deficient in an E. coli repair system - this system carries out homologous recombination, which can shuffle things around, which you might not want
- an endA mutation makes cells endonuclease I deficient - they don’t make a nonspecific endonuclease (nucleic acid cutter) that’s E. coli normally keep in their periplasmic space - this mutation can keep your plasmid from getting degraded
another thing to look for is the potential for blue-white screening: https://bit.ly/bluewhitescreening
some bacterial plasmids are designed for blue-white screening, including: pGEM-T, pUC18 and pUC19, & pBluescript
but the host cells need to be compatible with it too - some that are: XL1-Blue, DH5α, DH10B, JM109, STBL4, JM110, & Top10
When looking for a cDNA clone, the first place I normally look is Addgene. Although it sounds like it would be some commercial entity just out there to make a profit, it actually serves as a nonprofit plasmid repository - labs can send a sample of their plasmids to and Addgene will propagate them (make more copies) and distribute them to the public for a minimal fee ($75/plasmid when I just ordered some).
Addgene is a great place to start because, if authors have deposited the plasmids they used for protein expression, and that expression was in the system you want (e.g bacterial expression not a mammalian expression vector) then you won’t even have to subcclone! and the plasmid might be optimized for good expression - or at least you know it should work). If you can only find the gene cloned into a vector for a different expression system, don’t worry - you can just subcclone it into one you want - more work, but shouldn’t be an issue - and definitely not worth paying commercial companies an arm and a leg to get a version in the plasmid you want.
note: Addgene also has a really great educational blog - I suggest checking out their Plasmids 101 guide https://www.addgene.org/educational-r...
Another source which you can turn to if Addgene turns up short is DNASU, which is a depository that has plasmids containing cDNAs for “all” genes, even those that people haven’t worked with already. see comments for rest








Информация по комментариям в разработке