For more information, log on to-
http://shomusbiology.weebly.com/
Download the study materials here-
http://shomusbiology.weebly.com/bio-m...
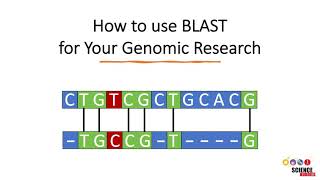
In bioinformatics, Basic Local Alignment Search Tool, or BLAST, is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences. A BLAST search enables a researcher to compare a query sequence with a library or database of sequences, and identify library sequences that resemble the query sequence above a certain threshold. Different types of BLASTs are available according to the query sequences. For example, following the discovery of a previously unknown gene in the mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. The BLAST program was designed by Stephen Altschul, Warren Gish, Webb Miller, Eugene Myers, and David J. Lipman at the NIH and was published in the Journal of Molecular Biology in 1990.
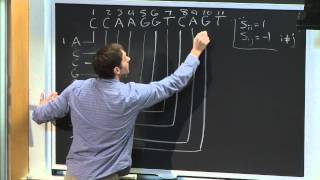
Using a heuristic method, BLAST finds similar sequences, not by comparing either sequence in its entirety, but rather by locating short matches between the two sequences. This process of finding initial words is called seeding. It is after this first match that BLAST begins to make local alignments. While attempting to find similarity in sequences, sets of common letters, known as words, are very important. For example, suppose that the sequence contains the following stretch of letters, GLKFA. If a BLASTp was being conducted under default conditions, the word size would be 3 letters. In this case, using the given stretch of letters, the searched words would be GLK, LKF, KFA. The heuristic algorithm of BLAST locates all common three-letter words between the sequence of interest and the hit sequence, or sequences, from the database. These results will then be used to build an alignment. After making words for the sequence of interest, neighborhood words are also assembled. These words must satisfy a requirement of having a score of at least the threshold T, when compared by using a scoring matrix. One commonly-used scoring matrix for BLASTp searches is BLOSUM62, although the optimal scoring matrix depends on sequence similarity. Once both words and neighborhood words are assembled and compiled, they are compared to the sequences in the database in order to find matches. The threshold score T determines whether or not a particular word will be included in the alignment. Once seeding has been conducted, the alignment, which is only 3 residues long, is extended in both directions by the algorithm used by BLAST. Each extension impacts the score of the alignment by either increasing or decreasing it. Should this score be higher than a pre-determined T, the alignment will be included in the results given by BLAST. However, should this score be lower than this pre-determined T, the alignment will cease to extend, preventing areas of poor alignment from being included in the BLAST results. Note, that increasing the T score limits the amount of space available to search, decreasing the number of neighborhood words, while at the same time speeding up the process of BLAST. Source of the article published in description is Wikipedia. I am sharing their material. Copyright by original content developers.
Link- http://en.wikipedia.org/wiki/Main_Page








Информация по комментариям в разработке