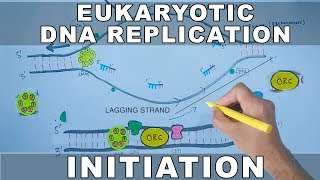
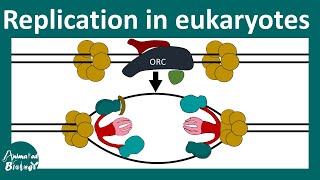
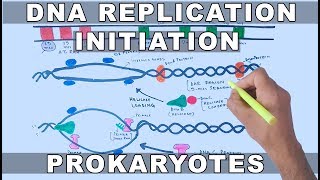
DNA Replication in Eukaryotes | Elongation
Скачать DNA Replication in Eukaryotes | Elongation бесплатно в качестве 4к (2к / 1080p)
У нас вы можете скачать бесплатно DNA Replication in Eukaryotes | Elongation или посмотреть видео с ютуба в максимальном доступном качестве.
Для скачивания выберите вариант из формы ниже:
Cкачать музыку DNA Replication in Eukaryotes | Elongation бесплатно в формате MP3:
Если иконки загрузки не отобразились, ПОЖАЛУЙСТА,
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если у вас возникли трудности с загрузкой, пожалуйста, свяжитесь с нами по контактам, указанным
в нижней части страницы.
Спасибо за использование сервиса video2dn.com







Информация по комментариям в разработке